
Image of the BugSigDB Homepage (https://bugsigdb.org/Main_Page)
Welcome back! It’s been an interesting couple of weeks as an Outreachy intern at Bioconductor. I’ve been working on the Microbiome Study Curation project which uses BugSigDB, by reviewing already curated studies. In this blog, we’ll explore BugSigDB, a powerful tool that unravels the mysteries of microbiomes.
The Bioconductor community

An image of Bioconductor landing page (https://www.bioconductor.org/)
Let us begin from the origin. BugSigDB is a project belonging to Bioconductor, a free and open source software community for bioinformatics. It is an amazing group of researchers, scientists, and developers who work together to develop and maintain open source software for bioinformatics and computational biology. The goal of the Bioconductor project is to develop, support and disseminate free open source software that facilitates reproducible analysis of data from current and emerging biological assays.
What is BugSigDB all about?
BugSigDB is a treasure trove of manually curated microbial signatures from published microbiome differential abundance studies, that are standardized to the NCBI taxonomy. These microbial signatures are specific combinations of microbial species that are indicative of a particular condition, environment or disease. The microbiome is simply a community of microorganisms colonizing a particular habitat. In humans, it describes the microorganisms living in or on a particular part of the body. The human microbiome is diverse, as there are as many microbes as there are human cells in the body (check out this study). Changes in the microbiome due to different factors including diet, medications, and environmental exposures can disrupt a person’s microbiome in ways that could increase the likelihood of developing conditions such as diabetes, obesity, cardiovascular and neurological diseases, allergies, and inflammatory bowel disease. It is important to note that the microbiome is not limited to humans. Animals, plants and even environments are also made up of this community of microorganisms. Differential abundance studies are studies that compare the relative abundance of different microbial species in different samples in conditions of interest. They help us to understand how microbial communities vary under different conditions. I explained what the term curation means in my previous blog. Alongside the microbial signatures, the host species, study design, statistical method, sampled body site, health outcomes, and other vital information are also curated. This makes the database information-filled, such that you can view the results of differential studies without necessarily opening the research paper. Its UI has several features including that make it beginner-friendly including but not limited to:
- the option to browse studies by different filters labelled 1 in the image below,
- the option to search for a particular taxa and view the studies it appears labelled 2 below, and
- the advanced search option labelled 3 below, which allows you to search for a taxa in a more streamlined fashion.
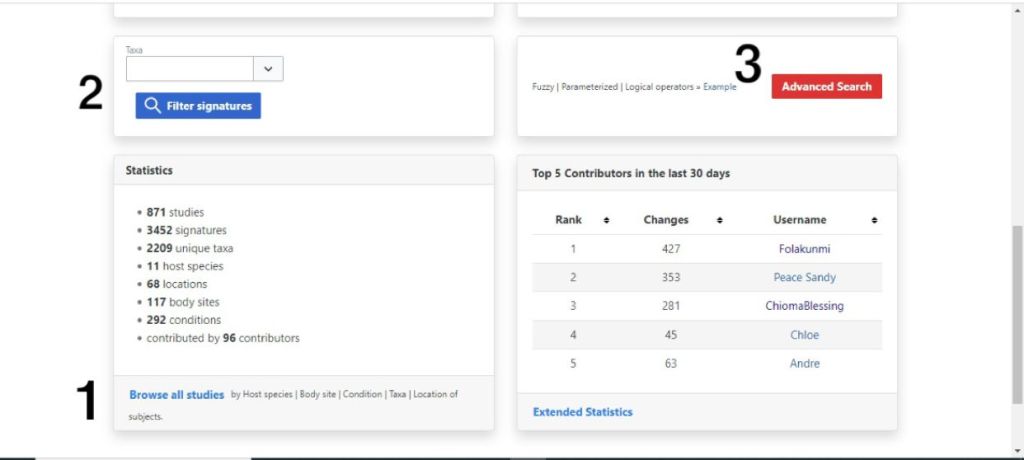
An image of BugSigDB home page (https://bugsigdb.org/Main_Page)
For in-depth knowledge on the technicalities and functions of BugSigDB, read this article by Geistlinger et al.
How to contribute to BugSigDB
Now that you know what BugSigDB is, let me show you how you can contribute to this database. There are many ways to contribute to BugSigDB, all of which can be seen on the help page. However, you are first required to get an understanding of the project by studying the onboarding materials;
- Read on the microbiome here; its general background, how differential abundance studies are done, statistical tests that are carried out, and other vital terminologies that you’ll come across during curation.
- Watch this collection of video tutorials which includes a video on the summary of what BugSigDB is, including an overview of the home page, a walk-through video of how to curate a new study, and a demo on how to work with BugSigDB data in R (for data analysts).
- To understand the guidelines of curating a study from scratch, read the Curation policy
- Request a bugSigDB account and wait for your temporary password. The temporary account email sometimes goes to your spam folder! On receiving the mail, change your password to one you’ll remember easily for subsequent logins.
- Join the #bugsigdb channel of the community-bioc Slack team
- Now, you are ready!
Welcome to BugSigDB!
Now, you are ready to start BugSigDB-ing by adding a new study and curating from scratch, or carrying out review tasks which include:
- helping to match unknown taxa names to the NCBI taxonomy. This is what we jokingly refer to as the “detective work” because it often involves rigorous internet searching. Watch a 2-hour video of Levi Waldron carrying out this task here;
- completing studies of interest that have been added to the database but have not yet been curated and are missing experiments and signatures here; and
- fixing improper values by matching study conditions to the Experimental Factor Ontology, body sites to the Uber-anatomy Ontology, and other vocabulary to BugSigDB-defined values.
View previously curated studies to get an inkling of what they look like by clicking the “random page” signified below.
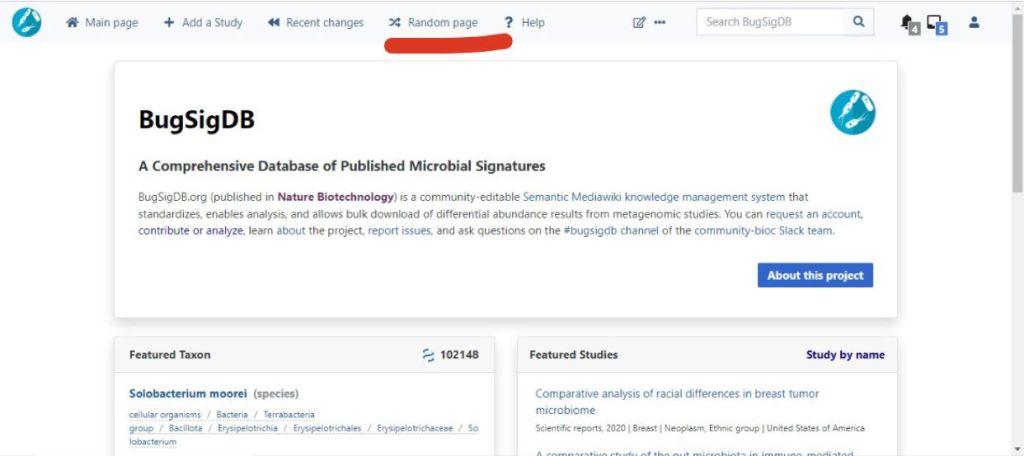
An image of BugSigDB home page (https://bugsigdb.org/Main_Page)
For Data Analysis
If you are a data analyst who’d love to analyze BugSigDB data; scroll down to the “Information for data analysts” part of the help page:
- watch these videos on BugSigDB use, search, and export and Importing BugSigDB Data in R, and
- scroll down to the “Information for data analysts” part of the help page.

An image of BugSigDB help page (https://bugsigdb.org/Help:Contents)
How to get help
The Biconductor community is a very warm community. I experienced the kindness of the community members (both applicants and mentors) right from the contribution phase of the Outreachy program. If you are ever confused or stuck, DO NOT HESITATE to ask for help on the #bugsigdb Slack channel. There are more than enough people willing to help you!
Welcome, and have a fun time BugSigDB-ing! 🥳🎉